Introduce some network models and their generating algorithms.
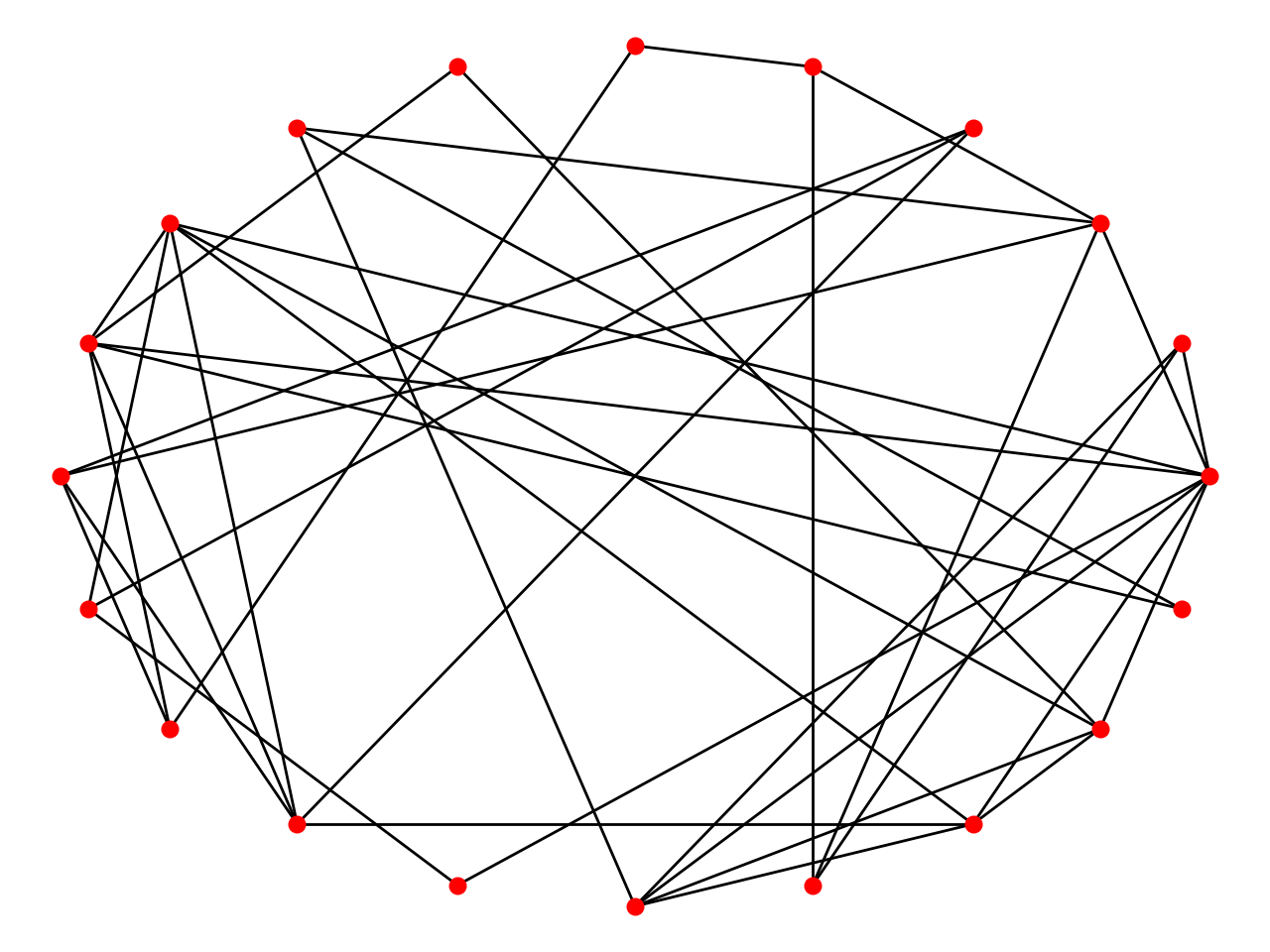
ER Model
The Erdős–Rényi model is either of two closely related models for generating random graphs.
In the model of Erdős and Rényi, all graphs on a fixed vertex set with a fixed number of edges are equally likely; in the model introduced by Gilbert, each edge has a fixed probability of being present or absent, independently of the other edges.
Algorithm
- Initialize adjacency matrix and probabilty p of linking.
- For all pairs of vertices, generating a random number~(0, 1). If this number is larger than p, add a edge between this pair of vertices.
- Do # step 2 until all vertices are choosed.
An efficient way to implement:
1 | def generateERGraph(graph_size, probability): |
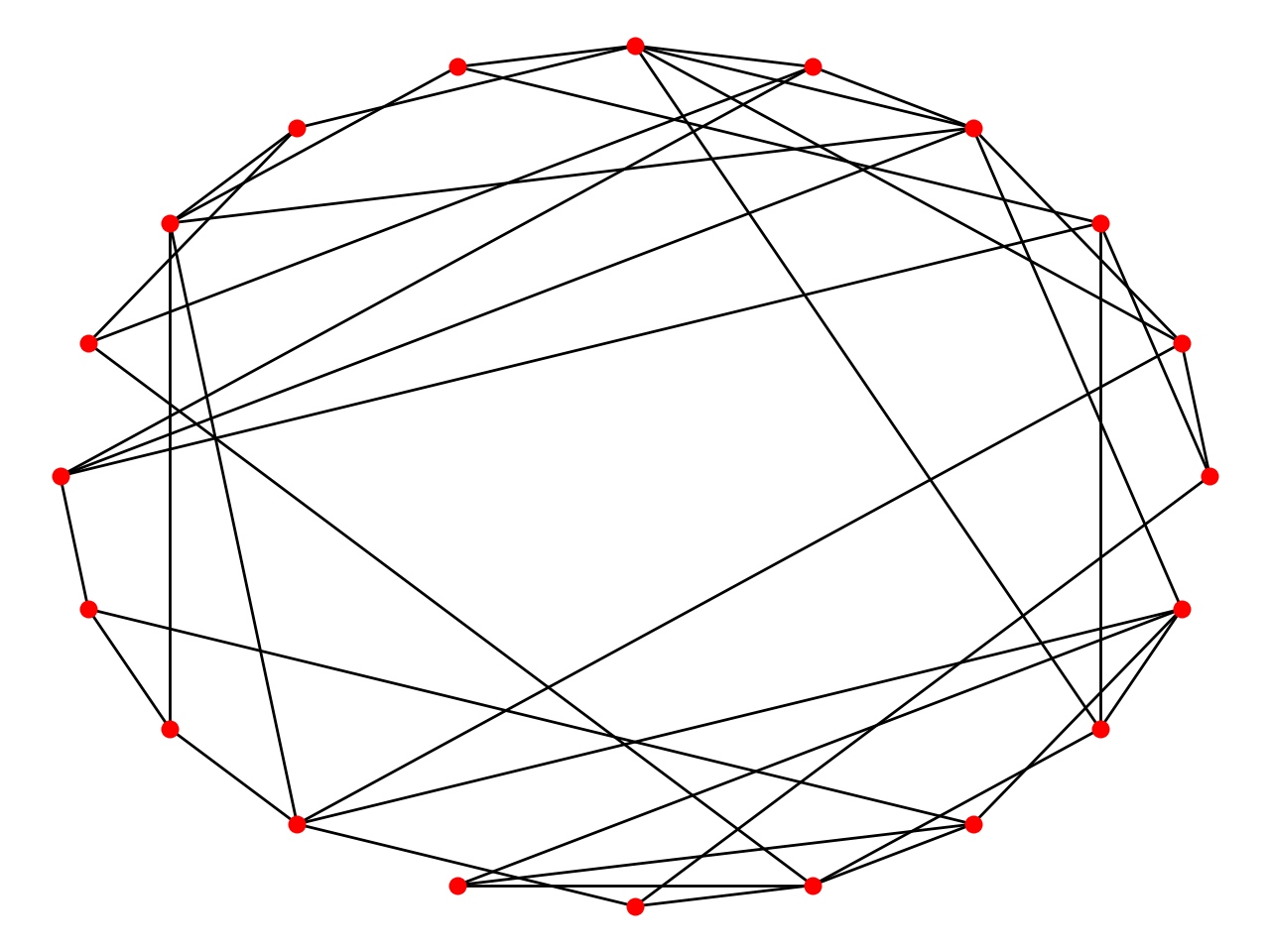
WS Small World Model
The Watts–Strogatz model is a random graph generation model that produces graphs with small-world properties, including short average path lengths and high clustering.
Algorithm
- Initialize adjacency matrix, number of edges with neighbors and probabilty p of linking.
- Connect each node to its right k/2 neighbors.
- For each edge u-v in ring, with p, randomly selecting existing node w and add new edge u-w.
1 | def generateSmallWorldGraph(graph_size, num_neighbors): |
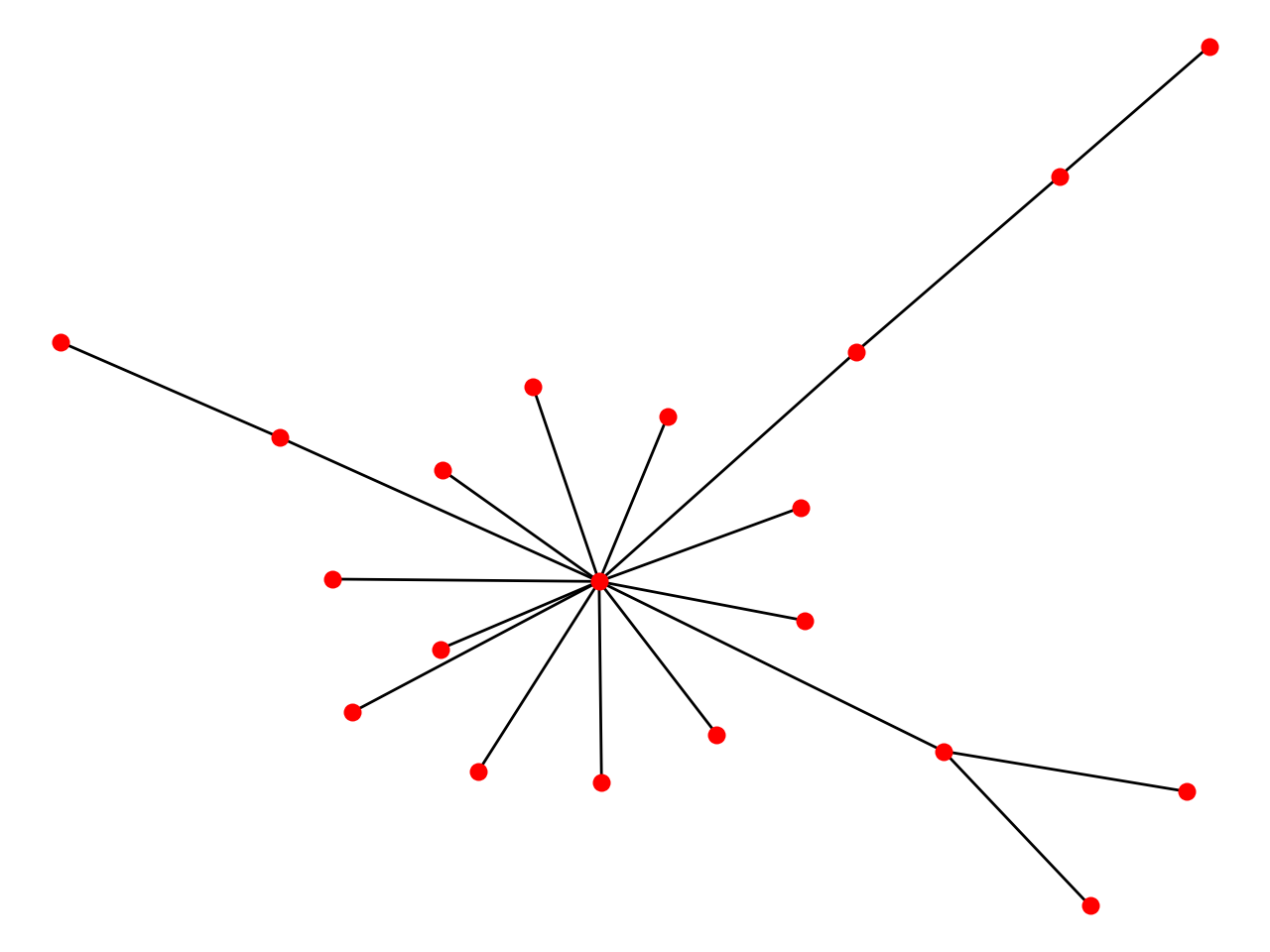
BA Model
The Barabási–Albert (BA) model is an algorithm for generating random scale-free networks using a preferential attachment mechanism.
Algorithm
- Initialize adjacency matrix and number of edges to attach from a new node to existing nodes. Choose a initial vertex v, a target list and a repeated list both with size m.
- Link v and vertices from target.
- Add [v]*m and all vertices from target to repeated list.
- Update target list by randomly choosing m vertices from repeated list and then choose a new vertex v to go # step 2.
1 | def generateBAGraph(graph_size, num_new_edges): |
